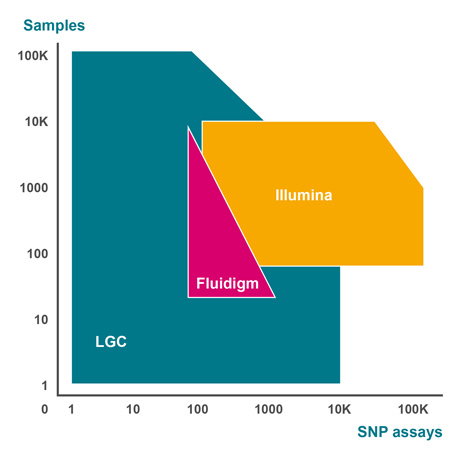
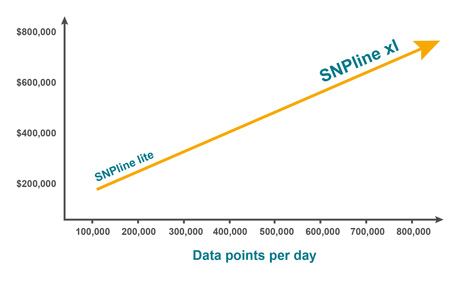
Developed and optimised over many years within our high-throughput genotyping service laboratories in Europe and North America, the SNPline™ workflow consists of all instruments, software, and validated procedures we use to establish a highly cost-effective, flexible, and scalable PCR-based genotyping solution. With an open-format design and modular framework, customers have the option of adopting one of two standard configurations: the SNPline Lite and SNPline XL, or they can build a custom configuration that incorporates equipment they already own.
Schematic illustration of the SNPline workflow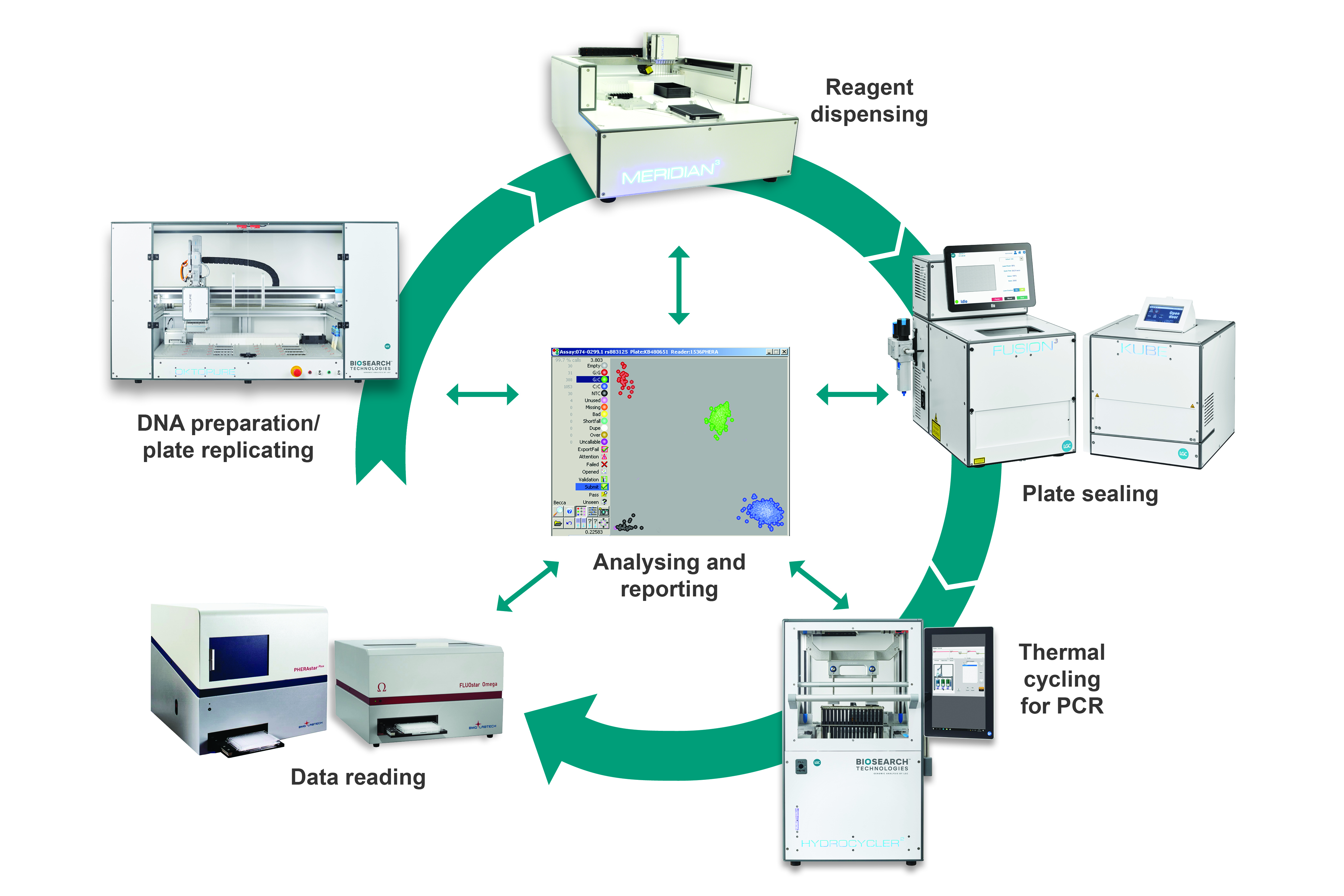
Originally developed to enable high throughput genotyping using our - KASP™ genotyping chemistry in the LGC's laboratories, the SNPline workflow is essentially a high throughput PCR workflow and, as such, it also lends itself to almost all high-throughput PCR applications, including other PCR-based genotyping methods, and high throughput expression profiling using RT-PCR.
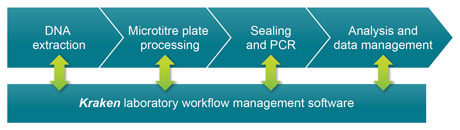
The workflow is comprised of discreet modular components and procedures that have been fully validated in our service laboratory environment, and that we continue to use extensively today. The process begins with DNA extraction from biological samples to obtain template DNA for downstream PCR applications. The DNA is typically extracted in plate format, and these DNA samples are subsequently arrayed into either 96-, 384- or 1536-well plates depending upon the scale of the project. The appropriate PCR reagents are dispensed into the DNA plates during the liquid dispensing step, and plates sealed in preparation for thermal cycling. Once the thermal cycling stage is complete, reaction plates can be read to obtain the raw data. Data analysis is then performed to enable final results to be reported.
Tying the process together into a complete workflow is Kraken™ workflow management software, which is an incredibly powerful tool that is integral to our entire service operation. Kraken takes control of sample management by tracking the storage locations of all sample plates and KASP Assay aliquots using 2D barcodes, and is also used for direct instrument control. Data analysis and reporting are also performed through Kraken, and this management software therefore provides a robust audit trail for the entire laboratory workflow.
We offer two standard configurations of the SNPline workflow:
- SNPline Lite for lower throughput applications (up to 20,000 data points per day; 96 and 384-well plates only)
- SNPline XL for higher throughput applications (up to 200,000 data points per day; 96, 384, and 1536-well plates).
Each configuration has the flexibility of being customised to best suit the needs of the customer, and the open-format workflow enables a customer’s existing instrumentation to be incorporated if appropriate.
The table below outlines the instruments and software that we recommend for each step within the SNPline Lite and SNPline XL workflows:
| Workflow step | SNPline Lite | SNPline XL |
|---|---|---|
| DNA extraction | oKtopure™ | oKtopure™ |
| Plate replication | Cyclops 2D reader + K-pette | 96 Rack High Speed 2D Reader + repliKator |
| Liquid dispensing | Multichannel pipettor | Meridian3™ dispenser |
| Plate sealing | Kube™ - thermal plate sealer | Fusion3™ - laser plate sealer |
| Thermal cycling | Hydrocycler2 ™ | Hydrocycler2 |
| Plate reading | BMG Omega F | BMG PHERAstar |
| Data analysis and reporting | KlusterCaller™ | Kraken™ |
Expert-level technical support
Our scientists have more than 10 years of genotyping experience using the SNPline workflow in our service laboratories. Our laboratory-based, scientific support team are on hand to provide full product and technical support. We can provide support for assays targeting difficult SNPs or InDels, and have extensive experience working with genomes from difficult organisms