Genomic selection: can it accelerate livestock breeding?
Livestock supports the livelihoods and food security of almost 1.3 billion people worldwide, contributing 40% of the global value of agricultural output. This makes the livestock sector one of the fastest growing parts of the agricultural economy, with opportunities for agricultural development, poverty reduction and food security gains. Increasing productivity throughout the whole sector will be essential to meet the growing demand for quality livestock products, whilst minimising its impact on the environment and the world’s natural resources.

Therefore, we need to focus on how to convert animal feed to meat protein more efficiently, looking towards advanced technologies and more efficient breeding methods to improve the genetic quality of livestock.
In this article, we investigate how genomic selection can now provide us with genomic information of increasing complexity, which can be utilised to evaluate polygenic traits such as feed conversion ratio, and how breeders and ranchers can leverage the latest genomic tools to meet the growing global demands on protein consumption.
Into the 21st century and beyond
The promise that more information at the DNA level would lead to more efficient genetic improvement than using only phenotypic records resulted in marker-assisted selection (MAS) which consists of two steps:
- mapping the genes underlying traits of interest: the quantitative trait loci (QTL);
- including that QTL information into the best linear unbiased prediction – an estimate of breeding value (BLUP-EBV).
We now apply complex statistical models to animal breeding programmes, through the estimation of (EBV) diversity evaluation of specific populations by application of BLUP linear modelling.1
Traditional MAS did not result in a widespread application of molecular markers in animal breeding beyond selecting for a few direct marker-to-phenotypic traits such as fat content in milk.2 This is due in part because many traits of interest in livestock production were much more complex, sometimes determined by hundreds of genes, with each eliciting small combinatorial effects on phenotype, making it nearly impossible to cost-effectively interrogate and make breeding decisions based on the complex and uncorrelated results.3
The breakthrough would come with the advent of whole-genome sequencing projects and made viable on a larger scale by next-generation sequencing (NGS). NGS enhanced the discovery of large numbers of single nucleotide polymorphisms (SNPs) that could be then associated with individual QTLs that could be identified as part of polygenic traits.
In contrast with MAS, which focuses on a small number of QTLs, and SNP markers associated with specific phenotypes, genomic selection (GS) uses a higher resolution set of markers from across the entire genome, assuring that all QTLs across the genome are in Linkage Disequilibrium (LD) (i.e. associated) with at least one SNP marker in close proximity.4
Ultimately this means that all the potential allelic effects that a specific SNP / set of SNPs might have on the phenotype can now be estimated at the level of the population rather than the individual. This enables breeders to draw cause-effect conclusions, based on statistical predictions, for these complex traits, and allow them to make informed decisions about which animals to breed to maximise the desired outcome, such as yield or protein conversion, and overall profitability of their herd or population.
What makes a cow a cow?
A major step towards improving the health and disease management of cattle and enhancing the nutritional value of beef and dairy products came with the sequencing of the bovine genome, in 2009.5 It contains 2.9 billion DNA base pairs, and 22,000 protein-coding genes, with the identification of two million SNPs. At the time, CSIRO’s Dr Ross Tellam predicted: “We could potentially achieve as much improvement in cattle breeding and production in 50 years as we have over the last 8,000 years of traditional farming.”

The Hereford breed was selected for the bulk of the project, and the sequence of the bovine's 29 chromosomes provides new insights into the bovine genome and the unique traits that could impact production. For example, chromosome breakpoints are rich in repetitive sequences, duplications and include specific variations in genes associated with lactation and immune response. For example, one of the changed genes, histatherin, produces a protein in cow's milk that has anti-microbial properties.
Cattle have one of the most highly rearranged genomes of all mammals, with more translocations and inversions of chromosome fragments, such as cats and even pigs, which are closely related to cattle.
Five of the 1,032 genes devoted to metabolic functions in humans are missing from the cattle genome or have radically diverged, suggesting that cattle have some unique metabolic pathways. At the time, Lewin quoted that these differences in metabolism, along with changes in genes devoted to reproduction, lactation and immunity are a big part of "what makes a cow a cow".
Genomic selection comes of age
Advances and cost reductions in NGS methods and bioinformatics and development of targeted genotyping by sequencing (GBS) methods allow us to efficiently and cost-effectively correlate whole sets of SNPs with desired phenotypes. The advancements in library preparation technologies and exponential drop in sequence data generation cost are propelling the routine use of NGS-based methods for large scale and high-throughput genomic selection in animal breeding programmes. Using an NGS approach helps bring traits or new varieties to market faster, lowers costs of breeding operations, and increases profitability.
LGC, Biosearch Technologies continues to play an ever-expanding role in partnership with our customers by applying cost-effective and fit-for-purpose targeted GBS service, SeqSNP, to enable our customers to advance their breeding programs to optimise health, welfare improve the economic value and quality of livestock products.
High-resolution sequence-based genotyping has been extensively applied to livestock breeding programs to accurately predict and select livestock where investment needs to deliver maximum genetic gains with each generation of offspring. This investment in genomic-based breeding decisions has improved the productivity of livestock breeding programs for complex traits, such as feed conversion ratio or feed efficiency. For example, the use of genomic selection in early measured traits for sheep and beef cattle produced 20-40% more genetic gain when combined with reproductive technologies.6
Targeted GBS is starting to displace arrays for routine screening in breeding programs. One such genotyping approach is the SeqSNP platform, developed at Biosearch Technologies. SeqSNP provides a cost-efficient, flexible and scalable mid-plex genotyping platform as a service, or as bespoke kits for in-house targeted GBS.
SeqSNP allows for the assessment of complex traits in all modern breeding programs and is ideally placed for the application of genomic selection. Independently analysed data substantiates that the SeqSNP service delivers genotyping data with high concordance to array genotyping and provides a more flexible and scalable alternative, without the minimum sample size commitments, and still achieving competitive pricing.
With SeqSNP, flexibility and conversion rates in probe library design are increased by the selection of sequences surrounding the target, rather than direct discrimination of the SNP, which also allows for accommodation of minor allele frequency in the surrounding sequence, and hence the identification of de novo SNPs.
An additional potential advantage of SeqSNP is that the use of two oligonucleotide probes for the sequencing of each given target provides not only cost-efficiency but also further confirmation of target alleles.
Figure 1 outlines the methodology used to produce probe libraries for targeted GBS in SeqSNP.
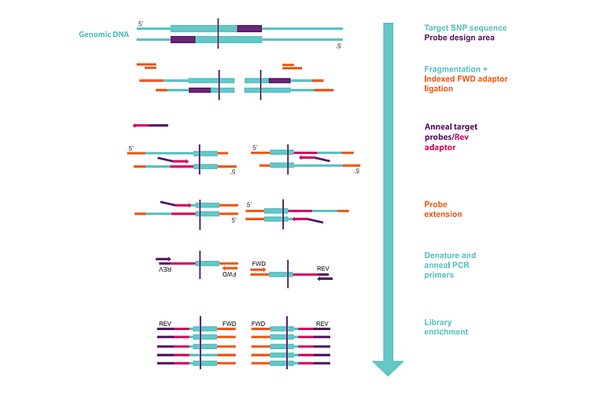
The advantages of using SeqSNP over other targeted GBS approaches include:
- Lower set-up costs, as costs are dependent on SNP and sample number, with no minimum sample commitments or up-front library development costs.
- Flexible marker selection: Up to 100K SNPs per sample in a single run. Ideally suited for 5,000 to 25,000 SNPs per sample.
- Flexible design in terms of non-fixed content: SNPs can be easily modified from run to run according to the traits examined, compared to fixed content arrays.
- Additional information gained on de novo variants (including structural variants) detected in target SNP region.
- Cost-effective: Highly efficient enrichment methods reduce day-to-day operation costs.
- Shorter development times for probe library production compared to array manufacture.
- Fragmentation of the gDNA replacing mechanical shearing means simultaneous digestion and labelling of DNA fragments simplify the workflow.
- Dual-index sample barcoding enables multiplex sequencing of over 3000 samples in a single sequencing lane, which allows for further scalability without limitations.
Perspectives and conclusions
The progress in sequencing technologies and their application to animal breeding and genomic selection seen over the last decade or so are game-changing but have still yet to realise their full potential.
Historically, the lack of genomic knowledge and access to sequencing technologies limited animal breeding programs to selection of key production traits that had a clearly defined genotype to phenotype correlations. The advancement of sequencing technologies, bioinformatics, and development of low-cost sequencing methods to interrogate a set of targeted SNPs, continues to expand the application of genomics to animal breeding programs.
These new methods add value to breeders and ranchers, who are now able to make informed decisions about mating choices, and take into account both key production and complex traits that improve overall value and profitability of their herds, and enable them to rise to the challenge that our increased protein diets place on our food supply.
This blog post was originally published on the LGC, Biosearch Technologies blog here.
REFERENCES
-
Fernando, R.F and Grossman, M (1989) Marker assisted selection using best linear unbiased prediction Genetics Selection Evolution, 21, 467.
-
Williamson, P. (2015) The Fat Controllers: Dairy Cattle Genetics and Milk Fat Composition, SPLASH! milk science update: May.
-
Goddard ME and Hayes BJ. (2009) Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet, 10: 381–391.
-
Hayes, B. and Goddard M. (2010) Genome-wide association and genomic selection in animal breeding, Genome, 53: 876-83.
-
Zimin, A.V., et al. (2009) A whole-genome assembly of the domestic cow, Bos Taurus Genome Biol., 10(4): R42.
-
Granleese, T., Clark, S. A., Andrew A. Swan A, A., Julius, H. J., Werf, V. (2015). Increased genetic gains in sheep, beef and dairy breeding programs from using female reproductive technologies combined with optimal contribution selection and genomic breeding values. Genet Sel Evol.; 47(1): 70. https://doi:10.1186/s12711-015-0151-3.
